Recent years have begun to close the gap regarding molecular mechanisms linking the gut microbiota to gastrointestinal (GI) diseases, including the inflammatory bowel diseases (IBD), types 1 and 2 diabetes, obesity and metabolic syndrome, and colorectal cancer. In some cases, mechanisms of action have been identified by which gut microbial organisms, their metabolic products, or other associated bioactive compounds perturb host immunity, often in the presence of pre-existing genetic risk factors. In a few cases, both live cell and microbially-derived small molecule interventions have proven to prevent or ameliorate GI inflammation, but to a large degree the therapeutic potential of targeted components of the microbiome has not yet been realized.
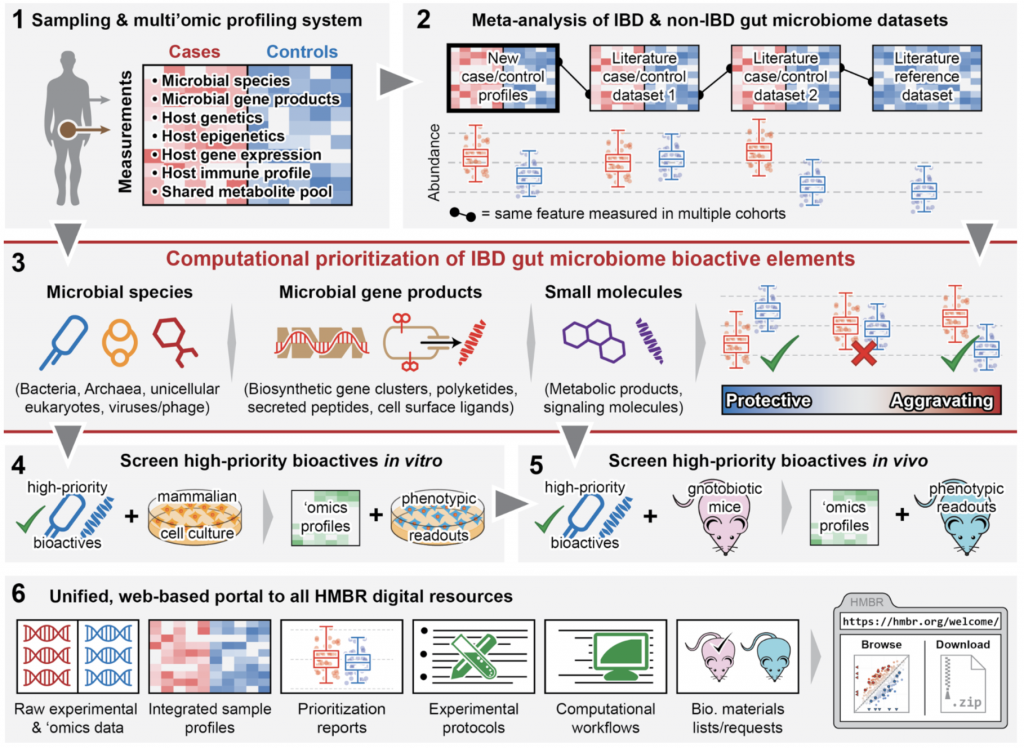
The Human Microbial Bioactives Resource (HMBR) will thus develop a comprehensive platform for discovery, validation, and early-stage translation of novel therapeutics derived from the microbiome. This resource will provide tools for 1) microbiome data generation, 2) microbiome meta-analysis, 3) bioactive lead prioritization, 4) in vitro screening, 5) in vivo screening in mouse models, and 6) an integrative online data, documentation, and training portal. The HMBR is being developed in response to the NIH NIDDK RFA DK-15-012, which calls for a, “community research resource of identified members of the microbiome and factors they elaborate which modulate human physiology or pathophysiology related to… digestive diseases.” As a driving biological problem for the system, we will also initially apply it to develop tunable modulators of the IL-10 and TNFα anti- and pro-inflammatory cytokine pathways implicated in IBD pathogenesis. Our aims for the HMBR thus entail:
- Providing an efficient end-to-end sampling and multi’omic profiling system for the host and microbiota in gastrointestinal disease.
- Integrating data from thousands of IBD microbiomes and tens of thousands of gut microbiome profiles, spanning dozens of datasets and meta-analyzing multiple countries, cohorts, and clinical centers.
- Computationally prioritizing potentially bioactive elements of the gut microbiome in IBD, including A) microbial species and strains (including viruses and eukaryotic microorganisms), B) microbial gene products (proteins, secreted peptides, biosynthetic gene clusters, etc.), and C) small molecules (e.g. metabolites or signaling molecules).
- Screening high-priority bioactives in vitro using mammalian cell and tissue models to characterize potential mechanisms of action and identify host-microbe-metabolite interactions.
- Phenotypic readouts (cell population, cellular function-based, and preclinical IBD models) for successful bioactives in conventional and gnotobiotic mice to assess activity in vivo.
- Providing all HMBR data, protocols, computational tools, microbial isolates, compounds, cell lines, mice, screening results, and training material to the community through a unified web-based portal.
- Exercising the resource with a driving biological application by verifying and characterizing novel modulators of the IL-10 anti-inflammatory and TNFα pro-inflammatory responses.
